KINtaro Database
FAMILY: HipA_C (PKLF000019)
Description
HipA kinases are crucial in the stress response mechanisms of E. coli and various other Gram-negative bacteria. They achieve this by triggering a dormant state known as persistence. In E. coli, HipA forms a toxin-antitoxin system alongside its neighboring genomic component, the HipB antitoxin. HipA phosphorylates glutamyl-tRNA synthetase, leading to the inhibition of protein synthesis and halting of growth. The presence of HipB inhibits the activity of HipA by binding to it, and HipB also acts as a transcriptional autosuppressor for the hipBA operon [PMID: 36526881]. Family includes cell translocating kinase (CtkA). The possible role of CtkA is bacterial persistence and escape from host innate immune responses by apoptosis of macrophages [PMID: 25172221].
Origin
phmmer: 0.0001;
database: nr;
sequence_cutoff: 100aa;
clustering: cdhit: 90%;
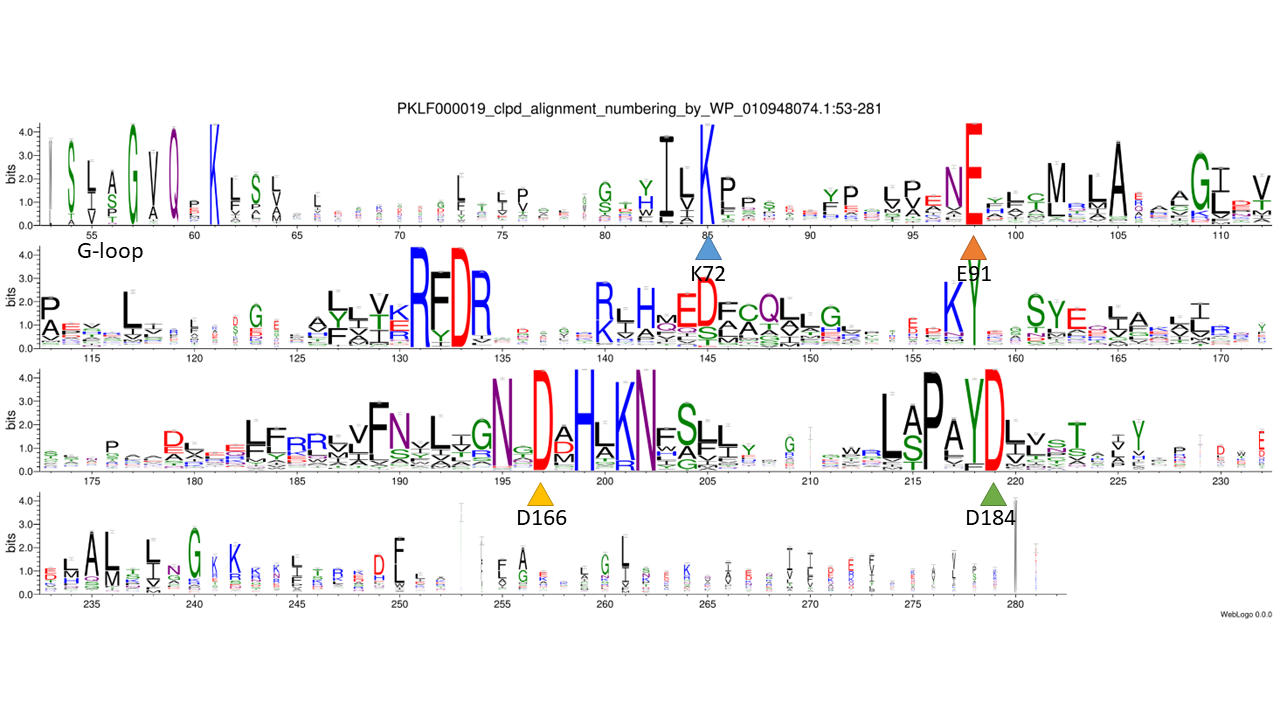
catalytic residues: based on - collapsed family logo, 3D structure model, PDB (3AKJ), FATCAT structural pairwise alignment with 3AKJ, 1ATP, 1O6Y and 6PWD, PMID: 21098302
Structure
PKL domain
HMM Model
WebLogo
 ×
×
![]()
Sequences
Aligned domain sequences
Download sequence
Unaligned domain sequences
Download sequence
Full sequences
Download sequence
Description
HipA kinases are crucial in the stress response mechanisms of E. coli and various other Gram-negative bacteria. They achieve this by triggering a dormant state known as persistence. In E. coli, HipA forms a toxin-antitoxin system alongside its neighboring genomic component, the HipB antitoxin. HipA phosphorylates glutamyl-tRNA synthetase, leading to the inhibition of protein synthesis and halting of growth. The presence of HipB inhibits the activity of HipA by binding to it, and HipB also acts as a transcriptional autosuppressor for the hipBA operon [PMID: 36526881]. Family includes cell translocating kinase (CtkA). The possible role of CtkA is bacterial persistence and escape from host innate immune responses by apoptosis of macrophages [PMID: 25172221].
Origin
phmmer: 0.0001; database: nr; sequence_cutoff: 100aa; clustering: cdhit: 90%; catalytic residues: based on - collapsed family logo, 3D structure model, PDB (3AKJ), FATCAT structural pairwise alignment with 3AKJ, 1ATP, 1O6Y and 6PWD, PMID: 21098302
Structure
PKL domain
HMM Model
WebLogo
